GEDI
The Global Ecosystem Dynamics Investigation (GEDI) mission is the first satellite with LiDAR survey capabilities that is designed for forest structure assessment. It provides high spatial resolution data with 25 m footprint diameter and spaced 60 m apart. The measurement lines are 600 m apart.
Data was processed into different levels, such as L2A to estimate canopy height and L4A showing ABG data with global-scale accuracy coverage.
In this development, data from the GEDIv002-L4A product, which has improved horizontal accuracy down to 10 meters, was used as a reference when developing the model (Ground Truth Reference).
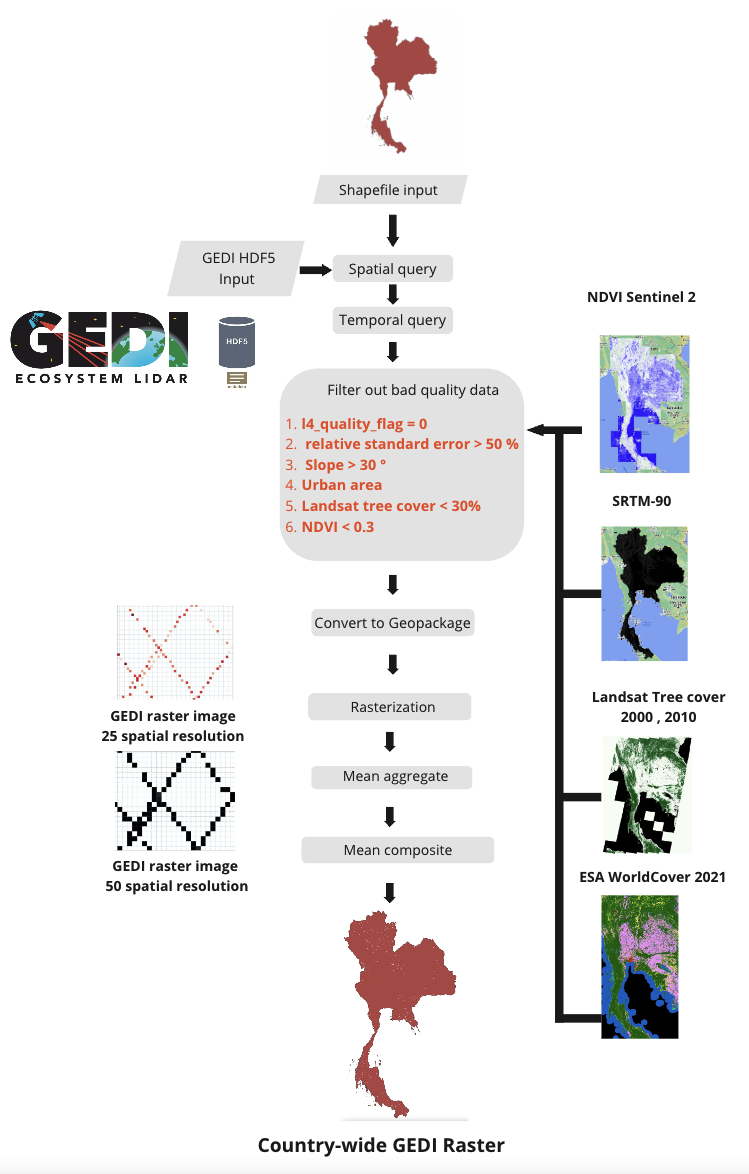
Because GEDI images are data obtained from reflectance of Earth’s surface to the sensor that are outside the atmosphere. There is interference from both internal factors from the equipment. and external factors from sunlight electromagnetic wave. Therefore, data that is subject to interference from various factors must be selected out in order to obtain the highest quality data before being processed to the model.
Data that does not meet the quality criteria. are follows:
Data not meeting the quality requirement of L4 level : parameter: ((agbd_se/agbd) x 100) > 50
Unreliable data with a relative standard error in AGB exceeding 50% : parameter: ((agbd_se/agbd) x 100) > 50
Data within sloped terrain areas exceeding 30%
- determined using NASA’s Shuttle Radar Topography Mission (SRTM) Digital Elevation Model (DEM) for slope calculations
- Data sourced from regions beyond the forested areas using WorldCover 2021 data from ESA, categorized into various types:
- Built-up : Class 50
- Sparse vegetation : Class 60
- Snow and Ice : : Class 70
- Permanent water bodies : : Class 80
- Herbaceous wetland : Class 90
- Moss and lichen : Class 10
Data in areas with a degraded forest landscape or sparse vegetation using EO tree cover data from Landsat satellites in 2000 and 2010 from NASA and filtering out areas with less than 30% forest cover,
Data in the area which has NDVI lower than 0.3
After filter out bad quality data. Convert HDF5 or .h5 format data to raster or grid data. (Rasterization) by giving the coordinates of each point as the center point (centroid) of the image pixel.
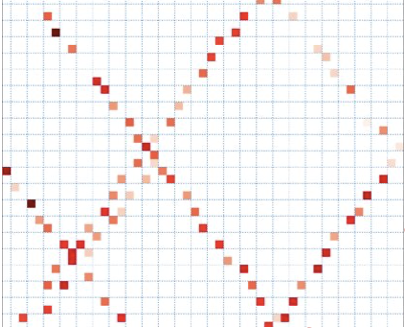
Then, GEDI data is obtained in the form of raster data with a spatial resolution of 25 meters according to the diameter of the data footprint. Example are show below.
Example of GEDI raster, 25 m spatial resolution.
Because the GEDI data has a horizontal geographic position accuracy of +-10 meters, errors may occur if the data is directly used with other earth observation data such as Sentinel-1 and Sentinel-2.
Aggregation is use to improve horizontal geolocation accuracy to data before importing into the model, resampling GEDI raster data to a spatial resolution of 50 meters, same as the Sentinel-1 and Sentinel-2 images
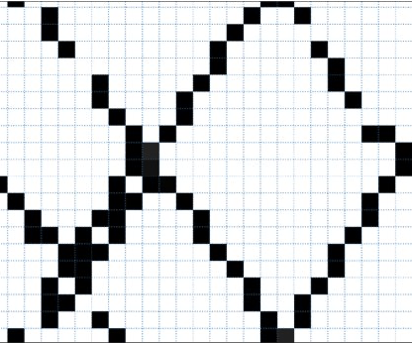
The procedures is done by calculating the average value of GEDI data that have more than 1 sample within the target pixel size of 50 m. Result example are show below.
Example of GEDI raster, 50 m spatial resolution.
All GEDI data obtained will be data from 2020-11-01 to 2021-04-30 based on Sentinel-1 and Sentinel-2 input data.
Time series data are composited by average data (Mean composite) with all images in the selected time period.
A pre-processing pipeline are show in diagram as follow.
Earth Engine Implementation
Example code snippet for preprocessing sentintel 1 and 2
- only requirement is to import target shapefile into Earth Engine environment and rename as target_shapefile
See documentation for how to upload various of assets into Earth Engine.
If you want to learn more about Earth Engine, explore End-to-End-Tutorial
Level 2
// import target shapefile in earth engine environment
var country = target_shapefile.geometry();
// define temporal range
var startDate = '2020-11-01'; //YYY-MM-DD
var endDate = '2021-04-30'; // YYY-MM-DD
////// Query Sentinel 2 to create NDVI image ////////////
// Country Name
var countryName = 'Thailand';
// Maximum cloud cover percentage
var cloudCoverPerc = 50;
// ----------------- SENTINEL-2 COLLECTION ------------------------
// We will use the Sentinel-2 Surface Reflection product.
// This dataset has already been atmospherically corrected
var s2 = ee.ImageCollection("COPERNICUS/S2_SR");
// Function to mask clouds S2
function maskS2srClouds(image) {
var qa = image.select('QA60');
// Bits 10 and 11 are clouds and cirrus, respectively.
var cloudBitMask = 1 << 10;
var cirrusBitMask = 1 << 11;
// Both flags should be set to zero, indicating clear conditions.
var mask = qa.bitwiseAnd(cloudBitMask).eq(0)
.and(qa.bitwiseAnd(cirrusBitMask).eq(0));
return image.updateMask(mask).divide(10000);
}
// Filter Sentinel-2 collection
var s2Filt = s2.filterBounds(country)
.filterDate(startDate,endDate)
.filterMetadata('CLOUDY_PIXEL_PERCENTAGE','less_than',cloudCoverPerc)
.map(maskS2srClouds);
///// NDVI /////
var addNDVI = function(image) {
var ndvi = image.normalizedDifference(['B5', 'B4']).rename('NDVI');
return image.addBands(ndvi);
};
var s2withNDVI = s2Filt.map(addNDVI);
// Max Annual NDVI
var s2MaxNDVIComposite = s2withNDVI.select('NDVI').max();
// NDVI mask
var NDVIMask03 = s2MaxNDVIComposite.lt(0.3);
var NDVIMask05 = s2MaxNDVIComposite.lt(0.5);
// ESA World Cover Filter
var ESA = ee.ImageCollection('ESA/WorldCover/v200').first();
var ESA_thai = ESA.clip(country);
// Create mask to filter GEDI
// Not equal 50 60 70 90 100
var shrub_land_mask = ESA_thai.eq(20);
var grass_land_mask = ESA_thai.eq(30);
var crop_land_mask = ESA_thai.eq(40);
var build_up_mask = ESA_thai.eq(50);
var sparse_veg_mask = ESA_thai.eq(60);
var snow_mask = ESA_thai.eq(70);
var water_mask = ESA_thai.eq(80);
var wetland_mask = ESA_thai.eq(90);
var moss_mask = ESA_thai.eq(100);
// Tree cover
var treecovdataset2000 = ee.ImageCollection('NASA/MEASURES/GFCC/TC/v3')
.filter(ee.Filter.date('2000-01-01', '2000-12-31'));
var treeCanopyCover2000 = treecovdataset2000.select('tree_canopy_cover');
var treeCanopyCover2000Mean = treeCanopyCover2000.mean();
var treecovdataset2015 = ee.ImageCollection('NASA/MEASURES/GFCC/TC/v3')
.filter(ee.Filter.date('2015-01-01', '2015-12-31'));
var treeCanopyCover2015 = treecovdataset2015.select('tree_canopy_cover');
var treeCanopyCover2015Mean = treeCanopyCover2015.mean();
// Tree Cover mask
var treeCanopyCover2000mask = treeCanopyCover2000Mean.lt(20);
var treeCanopyCover2015mask = treeCanopyCover2015Mean.lt(20);
var treeCanopyCover2000mask2 = treeCanopyCover2000Mean.gte(200);
var treeCanopyCover2015mask2 = treeCanopyCover2015Mean.gte(200);
// DEM
var SRTM = ee.Image('CGIAR/SRTM90_V4');
var elevation = SRTM.select('elevation').clip(country);
var slope = ee.Terrain.slope(elevation);
var slope_mask = slope.lt(30);
var qualityMask = function(im) {
return im.updateMask(im.select('quality_flag').eq(1))
.updateMask(im.select('degrade_flag').eq(0));
};
var slopeMask = function(im) {
return im.updateMask(slope_mask);
};
///// mask
/// Tree cover < 20
//1.Pixel is classify as water (ESA == 80)
var water_pixel = treeCanopyCover2000mask.and(treeCanopyCover2015mask).and(water_mask);
//2. Pixel locate in urban area and Maximum annual NDVI below 0.5
// 50 Built up
var buildup_pixel = treeCanopyCover2000mask.and(treeCanopyCover2015mask).and(build_up_mask).and(NDVIMask05);
// 60 Sparse Vegetation
var sparse_veg_pixel = treeCanopyCover2000mask.and(treeCanopyCover2015mask).and(sparse_veg_mask).and(NDVIMask05);
// 40 Crop land
var crop_land_pixel = treeCanopyCover2000mask.and(treeCanopyCover2015mask).and(crop_land_mask).and(NDVIMask05);
// 30 Grass land
var grass_land_pixel = treeCanopyCover2000mask.and(treeCanopyCover2015mask).and(grass_land_mask).and(NDVIMask05);
// 20 Shrub land
var shrub_land_pixel = treeCanopyCover2000mask.and(treeCanopyCover2015mask).and(shrub_land_mask).and(NDVIMask05);
// below 0.3 NDVI
var ndvipixel = treeCanopyCover2000mask.and(treeCanopyCover2015mask).and(NDVIMask03);
//Tree cover >= 200
//1.Pixel is classify as water (ESA == 80)
var water_pixel2 = treeCanopyCover2000mask2.and(treeCanopyCover2015mask2).and(water_mask);
//2. Pixel locate in urban area and Maximum annual NDVI below 0.5
// 50 Built up
var buildup_pixel2 = treeCanopyCover2000mask2.and(treeCanopyCover2015mask2).and(build_up_mask).and(NDVIMask05);
// 60 Sparse Vegetation
var sparse_veg_pixel2 = treeCanopyCover2000mask2.and(treeCanopyCover2015mask2).and(sparse_veg_mask).and(NDVIMask05);
// 40 Crop land
var crop_land_pixel2 = treeCanopyCover2000mask2.and(treeCanopyCover2015mask2).and(crop_land_mask).and(NDVIMask05);
// 30 Grass land
var grass_land_pixel2 = treeCanopyCover2000mask2.and(treeCanopyCover2015mask2).and(grass_land_mask).and(NDVIMask05);
// 20 Shrub land
var shrub_land_pixel2 = treeCanopyCover2000mask2.and(treeCanopyCover2015mask2).and(shrub_land_mask).and(NDVIMask05);
// below 0.3 NDVI
var ndvipixel2 = treeCanopyCover2000mask2.and(treeCanopyCover2015mask2).and(NDVIMask03);
var mapNonCanopyValue = function(image) {
var remapimage = image.where(image.select('rh95').lt(3) , 0.0)
.where(water_pixel , 0.0)
.where(buildup_pixel , 0.0)
.where(sparse_veg_pixel , 0.0)
.where(crop_land_pixel , 0.0)
.where(grass_land_pixel , 0.0)
.where(shrub_land_pixel , 0.0)
.where(ndvipixel , 0.0)
.where(water_pixel2 , 0.0)
.where(buildup_pixel2 , 0.0)
.where(sparse_veg_pixel2 , 0.0)
.where(crop_land_pixel2 , 0.0)
.where(grass_land_pixel2 , 0.0)
.where(shrub_land_pixel2 , 0.0)
.where(ndvipixel2 , 0.0);
return remapimage;
};
var dataset = ee.ImageCollection('LARSE/GEDI/GEDI02_A_002_MONTHLY')
.filterBounds(country)
.filterDate(startDate,endDate)
.map(qualityMask)
.select('rh95')
.map(slopeMask)
.map(mapNonCanopyValue);
var gedi_all = dataset.mean();
Map.addLayer(gedi_all, {min: 0, max: 100}, 'Gedi L2A', true);
// Export to cloud storage
Export.image.toCloudStorage({
image: gedi_all.toDouble(),
description: 'GEDI_L2A_32647_10m',
bucket: 'varuna-data-nonprod-analytic',
fileNamePrefix: 'biomass-estimation-project/vm-backup/Canopy_model_data/GEDI_L2A/GEDI_L2A_32647_10m',
scale: 10,
region: country,
maxPixels: 1e13
});
Export.image.toCloudStorage({
image: ESA_thai,
description: 'ESA_thai_10m',
bucket: 'varuna-data-nonprod-analytic',
fileNamePrefix: 'biomass-estimation-project/vm-backup/Canopy_model_data/ESA/ESA_thai_10m',
scale: 10,
region: country,
maxPixels: 1e13
});
Level 4
// import target shapefile in earth engine environment
var country = target_shapefile.geometry();
// define temporal range
var startDate = '2021-11-01'; //YYY-MM-DD
var endDate = '2022-04-30'; // YYY-MM-DD
// ESA World Cover Filter
var ESA = ee.ImageCollection('ESA/WorldCover/v200').first();
var ESA_thai = ESA.clip(country)
// Classification Mask
var build_up_mask = ESA_thai.neq(50);
var sparse_veg_mask = ESA_thai.neq(60);
var snow_mask = ESA_thai.neq(70);
var water_mask = ESA_thai.neq(80);
var wetland_mask = ESA_thai.neq(90);
var moss_mask = ESA_thai.neq(100);
var all_mask = build_up_mask.and(sparse_veg_mask).and(snow_mask).and(water_mask).and(wetland_mask).and(moss_mask)
var esaMask = function(im) {
return im.updateMask(all_mask);
};
// DEM -> Slope Mask
var SRTM = ee.Image('CGIAR/SRTM90_V4');
var elevation = SRTM.select('elevation').clip(country);
var slope = ee.Terrain.slope(elevation);
var slope_mask = slope.lt(30);
var slopeMask = function(im) {
return im.updateMask(slope_mask);
};
// GEDI Qualityi filter Mask
var qualityMask = function(im) {
return im.updateMask(im.select('l4_quality_flag').eq(1))
.updateMask(im.select('degrade_flag').eq(0))
.updateMask(im.select('agbd_se').divide(im.select('agbd')).multiply(100).lt(50));
};
// Dataset query
var dataset = ee.ImageCollection('LARSE/GEDI/GEDI04_A_002_MONTHLY')
.filterBounds(country)
.filterDate(startDate,endDate)
.map(qualityMask)
.map(esaMask)
.map(slopeMask)
.select('agbd');
// mean composite
var gedi_all = dataset.mean();
// Export data to gcs
Export.image.toCloudStorage({
image: gedi_all,
description: 'GEDI_L4A_25m_2021_2022',
bucket: 'varuna-data-nonprod-analytic',
fileNamePrefix: 'biomass-estimation-project/vm-backup/AGB_model_data/GEDI_L4A_25m_2021_2022',
scale: 25,
region: country,
maxPixels: 1e13
});